Abstract
Background: The implementation of genomic data from myelodysplastic syndrome (MDS) patients into clinical practice requires its association with MDS disease activity. However, the determination of a genotype-phenotype correlation in MDS where the disease phenotype is ineffective hematopoiesis, is not straightforward. Part of the problem may be that disease activity reflected by a change in blast count, a change in a peripheral blood count, or the acquisition of a new cytogenetic abnormality does not currently provide any information about the likely dynamics of hematopoietic progenitors underlying this change. Being able to reliably infer a change in the early hematopoietic progenitor compartment of MDS patients where the disease clone likely resides that can account for the phenotype of ineffective hematopoiesis is an important step in the development of a tool to measure disease activity over time.
We aim to apply a quantitative model of hematopoiesis based on parameters known to affect hematopoietic progenitor population dynamics, including rate of self-renewal, rate of proliferation, rate of differentiation, and rate of apoptosis. The mathematical formulation based on delay-differential equations has been successfully used to model the peripheral blood counts of CML and cyclic neutropenia. The goal is to identify unique MDS disease states based on the parameters described in the model, and to correlate these with both genotype and clinical outcomes.
Methods: MDS patients with IPSS intermediate-2/high diagnosed during the period 2010-2017 at the London Health Science Centre had their bone marrow aspirate and biopsy data at diagnosis, laboratory data while on treatment with azacitidine and following disease progression, and transfusion history collected. The time-dependence of the peripheral blood counts for each patient were modeled using a delay-differential equation (Colijn and Mackey, 2005), with parameters representative of rates of proliferation, self-renewal, differentiation, and apoptosis of the stem cell and progenitor compartments. The model was integrated with MATLAB's dde23 routine and the model parameters were fit to the peripheral blood counts using gradient descent for the constrained non-linear least squares problem. The patients were separated into two clusters using the k-means clustering of all the model parameters using the city-block distance measure. The number of clusters were determined using the Calinski-Harabasz criterion.
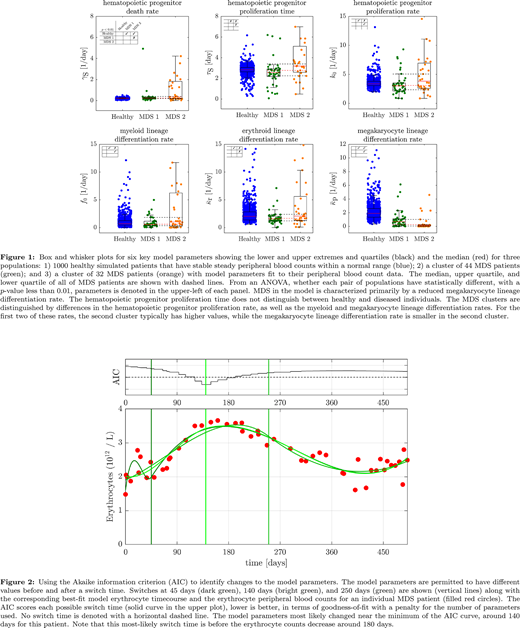
Results: Seventy-seven patients with a diagnosis of IPSS intermediate-2/high risk MDS were included in the analysis. Model fitting of the peripheral blood count data of 1000 simulated healthy patients whose blood count parameters were within their respective normal ranges over time yielded baseline variability of hematopoietic kinetic parameters (Figure 1). Two main groups of MDS patients could be identified from model fitting: MDS 1 and MDS 2, both of which could be best distinguished from the simulated healthy patients based on the parameter for the rate of megakaryocytic differentiation. MDS 1 and MDS 2 also had distinct parameters for the rates of hematopoietic progenitor proliferation, describing two different phenotypes of disease activity. The red blood cell count of a representative patient in MDS 1 is shown in Figure 2, where application of the Akaike information criterion identified the most likely time point when the hematopoietic kinetic parameters accounting for the peripheral red blood cell count changed after initiation of azacitidine. This time point preceded proven disease progression by 40 days.
Conclusion: Parameterization of hematopoiesis with a model that can infer the kinetics of hematopoietic compartments in the bone marrow provides a powerful tool for measuring ineffective hematopoiesis in MDS patients. The resulting kinetic parameters can identify distinct groups of MDS patients based on the phenotype of their disease. The next step would be to correlate the different kinetic parameters distinguishing these groups to their disease genotypes using next-generation sequencing. On an individual patient level, these parameters can identify the most likely time when the disease phenotype changes. The development of a dynamic score predicting the change in disease activity based on hematopoietic kinetic parameters derived from peripheral blood count data over time is ongoing.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal